A Boundless Platform
Plexium has developed comprehensive approach toward Targeted Protein Degradation, powered by our proprietary best-in-class platform, which enables us to discover a wide variety of TPD modalities, from molecular glues to monovalent degraders, while also identifying novel E3 ligases. This launches us toward the discovery of next-generation TPD drugs across multiple therapeutic areas. And, given the vast undrugged landscape of opportunity that remains, we’re just getting started.

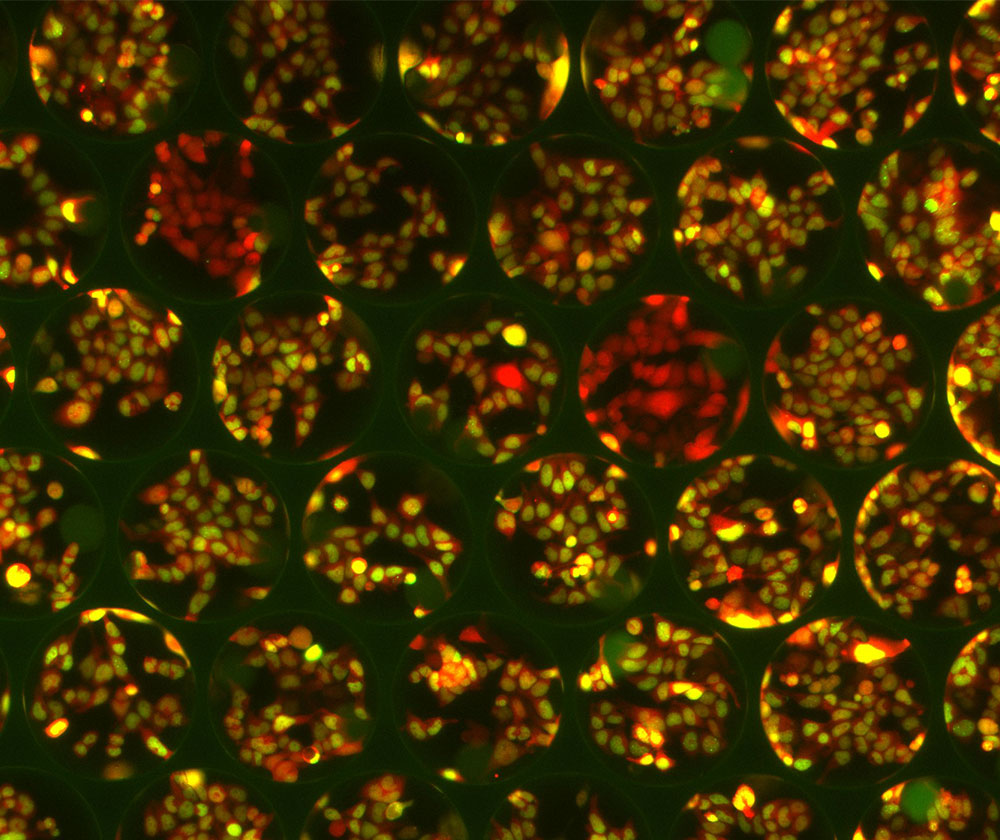
The Plexium Platform combines screening of purposefully designed “on-bead” DNA encoded libraries in ultra-high-throughput miniaturized cell-based assays that directly measure degradation of one or more proteins of interest in a disease-relevant cellular context. Plexium is utilizing its best in class, next generation platform to identify monovalent direct degraders or molecular glue degraders targeting pathogenic proteins.
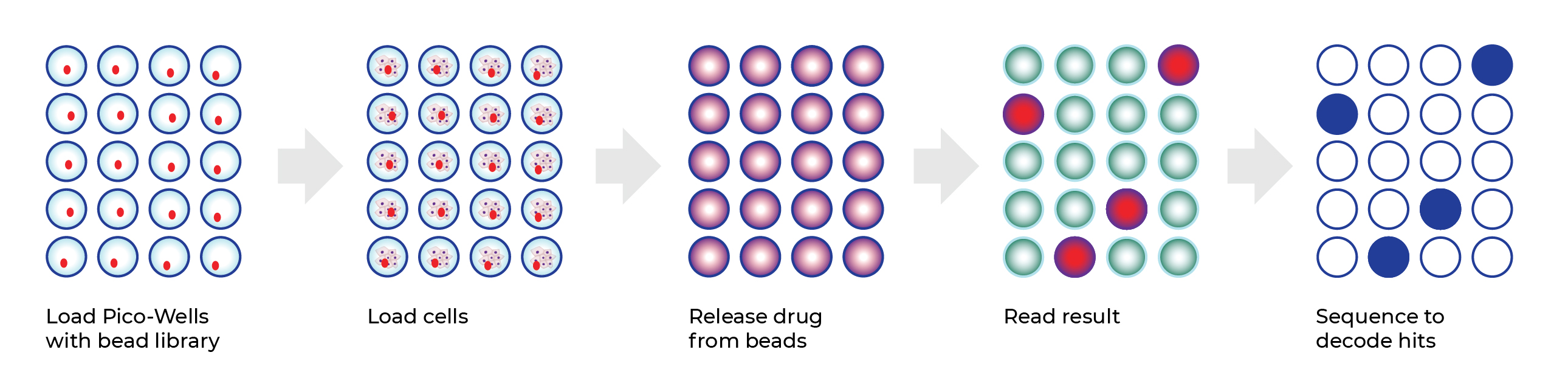
Advantages of the Plexium Platform include:
- Next-generation DEL combinatorial library approach identifies cell-active degrader drug candidates
- Primary assay readout is the identification of compounds that degrade a target of interest in representative cells in a disease-relevant cellular context
- A single approach can be used to identify molecular glues or direct monovalent degraders of pathogenic proteins
- Allows the full range of E3 ligases within a cell to potentially be accessed